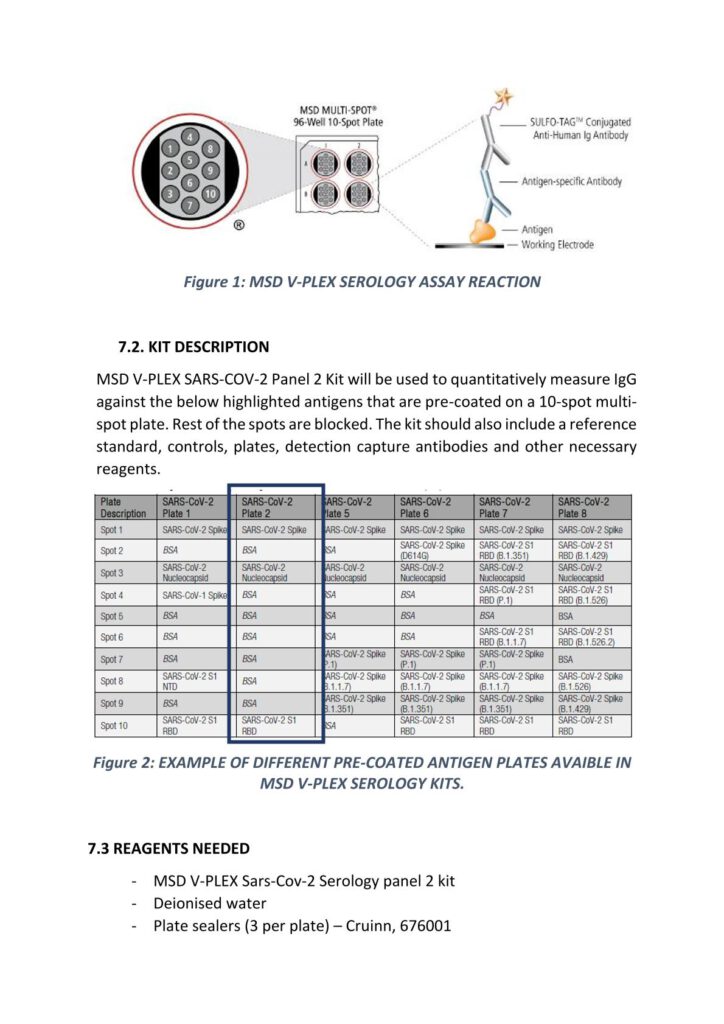
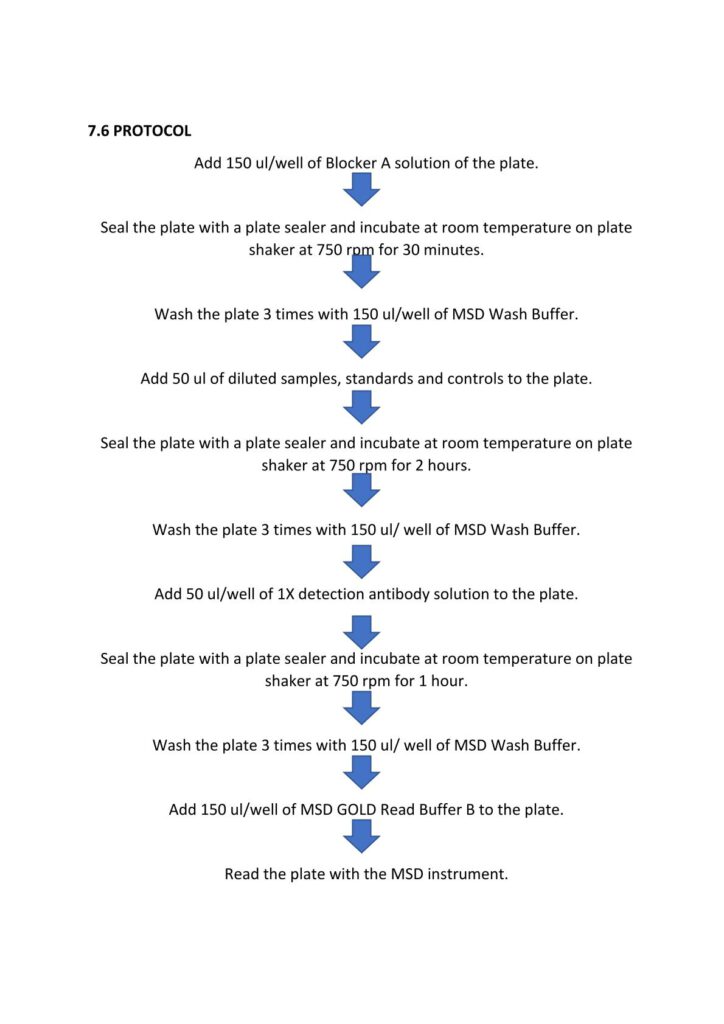
Immune Monitoring


Work package objectives:
WP8 will focus on compiling, and establishing a comprehensive catalogue of standardised immunological and genetic assays for COVID-19 vaccines that can be rolled out across the network. This enables a direct comparison of results from different laboratories which in turn facilitates the design and implementation of multi-site clinical trials. The catalogue will include standardised assay protocols for humoral, cellular, mucosal immune monitoring, and assays for genetic variants. The team aims to align and standardise protocols, reagents, and standards (such as international antibody standards from NIBCS) with other major initiatives and networks such as CEPI’s consortium of centralised COVID-19 labs, WHO’s ACT Accelerator and NIH’s Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV). The team will also review available scientific data to identify and prioritise additional immune monitoring assays that should be developed in the future.
WP Leads:
Prof. Jean-Daniel Lelièvre
Institut national de la santé et de la recherche médicale (INSERM), France
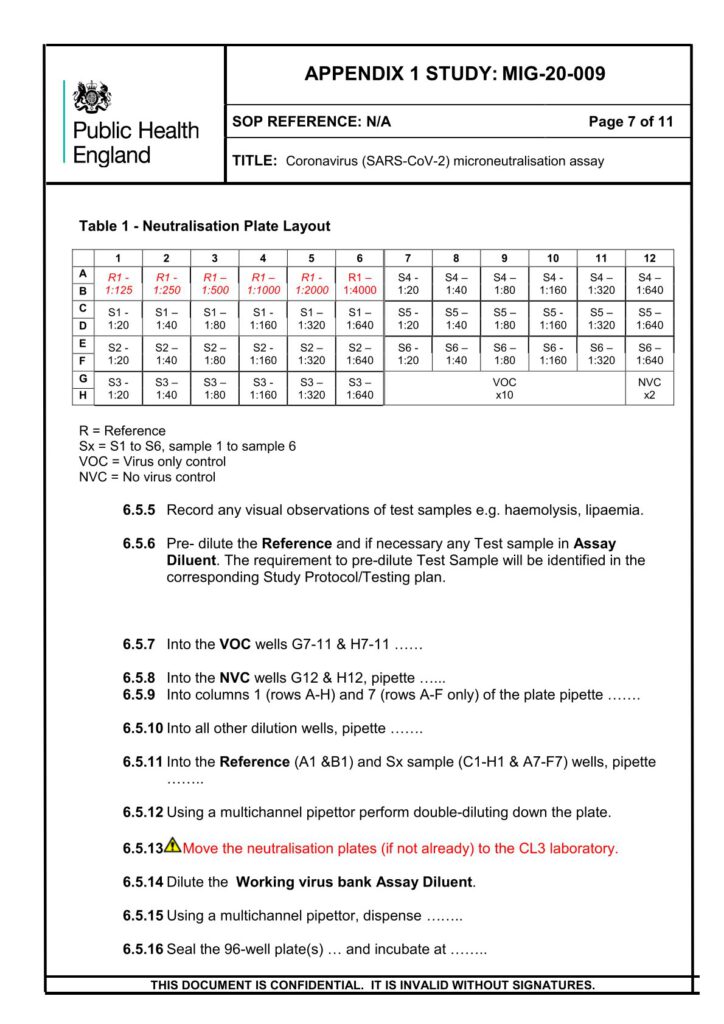
Prof. Ole F. Olesen
European Vaccine Initiative (EVI), Germany
Work Package Results and Protocols
All public documents can be shared upon request. Please contact: info@vaccelerate.eu.
Flow Cytometry based-Live SARS-CoV-2 Micro-Neutralisation assay
Dr. Virginie Gautier, Associate professor in Virology, Centre for Experimental Pathogen Host Research (CEPHR), School of Medicine, University College Dublin (UCD), Ireland
Protocol for Cellular Immune Response Assays (Institute of Health Carlos III)
Dr. Jordi Ochando
Protocol for Humoral Immune Response Assays (University of Bergen)
Prof. Rebecca Jane Cox Brokstad
Protocol for the Collection and Handling of Biological Materials
WP4
Anti-Sars-Cov-2 (Spike, Nucleocapsid and RBD) IgG Protocol for MSD Platform
Alejandro Abner Garcia Leon (UCD Dublin School of Medicine)
Protocol on the Preparation of PBMCs
WP4
Protocol on Whole Blood Processing
(WP4)
Saliva SARS-CoV-2 Antibody Analysis
Salivary Sample Collection
Salivary Sample Collection
CEPI SOPs
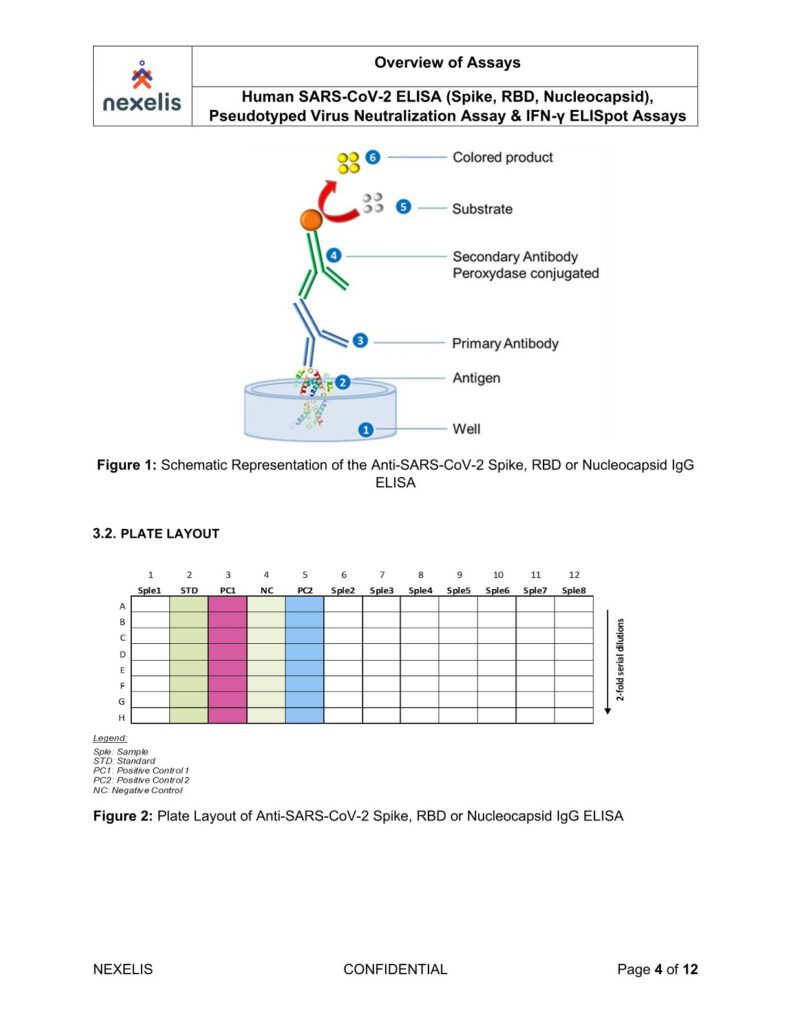
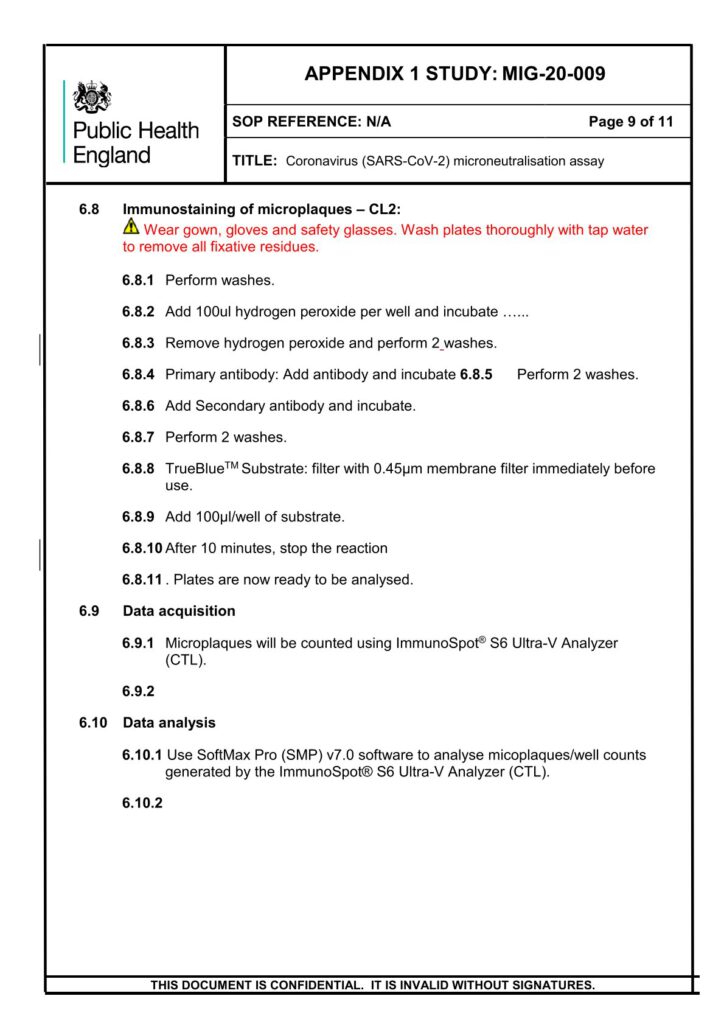
The human SARS-CoV-2 IgG ELISA is an indirect ELISA which is based on the antibody/antigen interactions. This ELISA allows the detection of SARS-CoV-2 Spike, RBD or Nucleocapsid specific IgG antibodies in human serum samples, and it can be adapted to animal samples as well.
The SARS-CoV-2 pre-fusion spike, RBD or Nucleocapsid recombinant antigen is adsorbed onto the 96-well microplate. Following incubation, the microplate is washed to remove unbound antigen and blocked to prevent non-specific binding. Standard, controls and sample dilutions are incubated in the coated microplate in which anti-SARS-CoV-2 Spike, RBD or Nucleocapsid IgG specific antibodies (primary antibodies) bind to the coated antigen.
Following incubation, the microplate is washed to remove unbound primary antibodies. Primary antibodies are detected with the addition of the anti-human IgG antibody (secondary antibody) conjugated to peroxidase. After incubation, the microplate is washed to remove unbound secondary antibodies.
The peroxidase substrate solution, tetramethylbenzidine (TMB), is added to the microplate and a colored product is developed which is proportional to the amount of SARS-CoV-2 Spike, RBD or Nucleocapsid IgG antibodies present in the serum sample. 2N H2SO4 is then added to stop the colorimetric reaction.
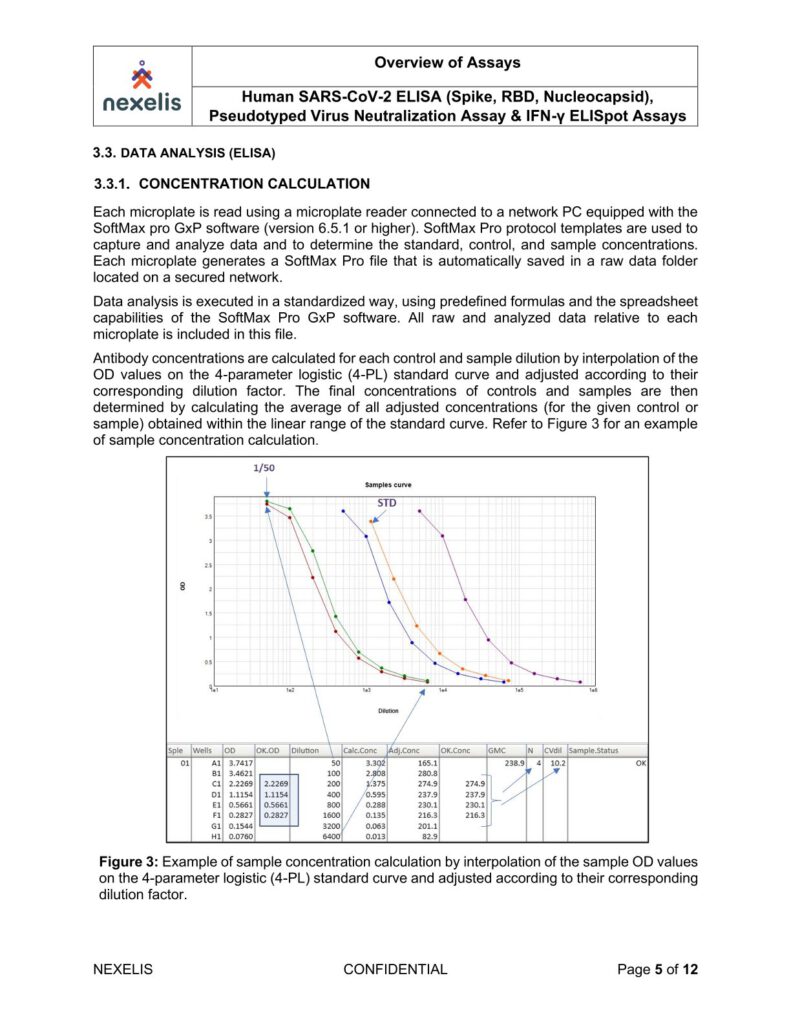
The absorbance of each well is measured using a microplate spectrophotometer reader at a specific wavelength (450/620 nm). A standard on each tested plate is used to calculate the amount of antibodies against SARS-CoV-2 Spike, RBD or Nucleocapsid in the sample according to the unit assigned by the standard (ELU/mL).
A peptide pool covering the whole S protein sequence (15-mer sequences with 11aa overlap) is prepared
in solutions at two times final concentration and added to a 96 well plate, 100μl/well. PBMC concentration
is adjusted to desired concentration, e.g.: 3 million/ml corresponding to 300,000 cells/well) and plated,
100μl/well using large orifice tips. The plates are incubated for 48-72 hours in a 37°C humidified incubator,
9% CO2.
Plates are washed three times with 0.05% Tween-PBS, 200μl/well and anti-human IFN-γ/IL-5 Detection
Solution is added (80μl/well). Plates are incubated at room temperature, two hours.
Plates are washed three times with 0.05% Tween-PBS, 200μl/well and the Tertiary Solution is added
(80μl/well). Plates are incubated at room temperature, two hours.
Plates are washed two times with 0.05% Tween-PBS, and then two times with distilled water, 200μl/well
each time before adding the Developer Solution (IFNy), 80μl/well. Plates are incubated at room
temperature, 15 minutes. The same steps are repeated for the second Developer Solution (IL-5).
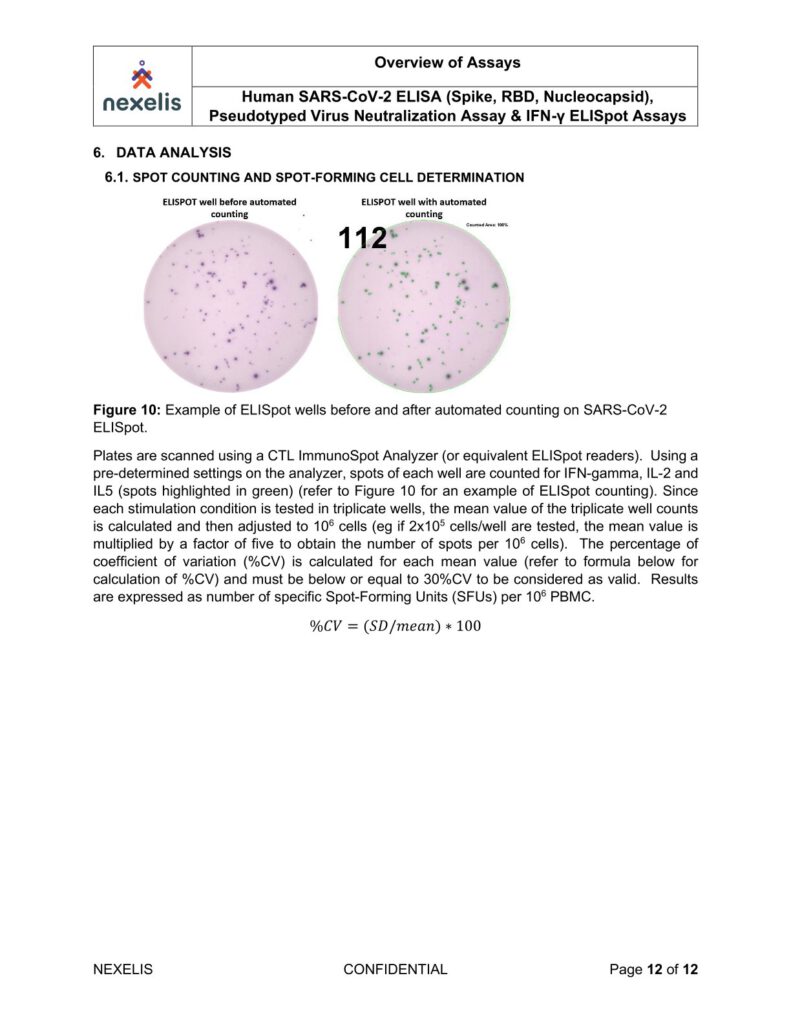
Plates are air-dried for two hours in running laminar flow hood or for 24 hours face down on paper towels
on bench top and then scanned and counted with an Immunospot analyzer.
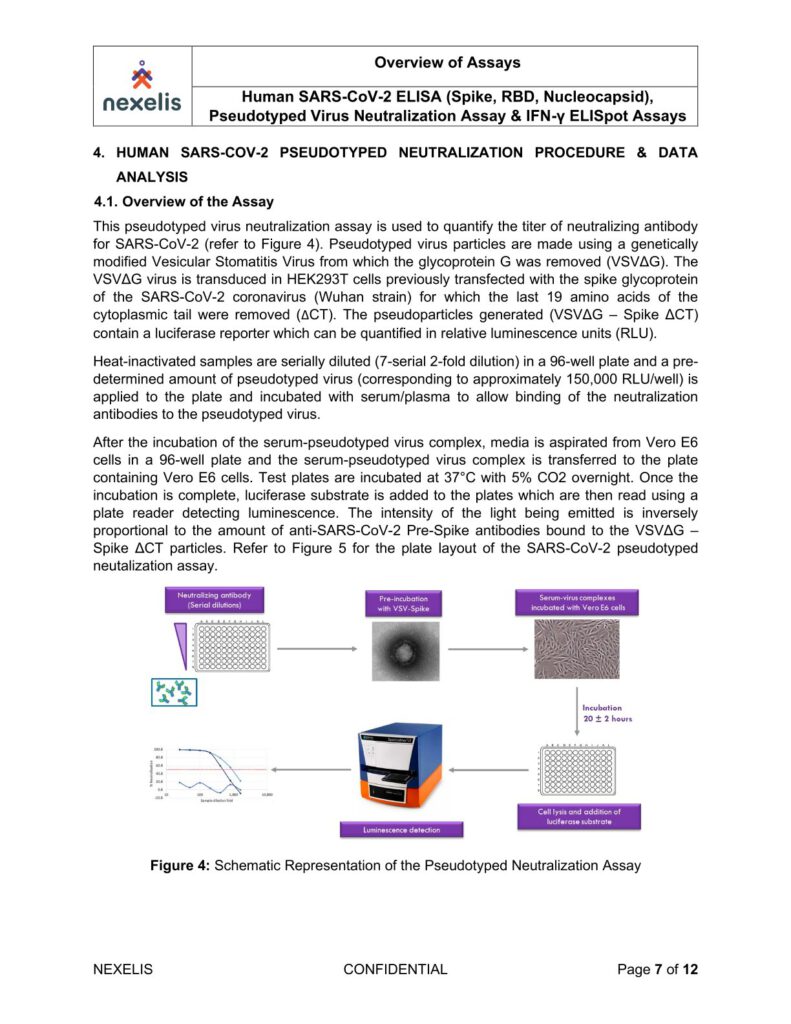
Heat-inactivated (56°C for 30 min) serum samples are serially diluted and incubated with approximately
60 PFU of wild type SARS-CoV-2 (2019-nCoV/Victoria/1/2020), for 1 h at 37°C in 5% CO2. Samples are
incubated with Vero E6 monolayers in 96-well plates under MEM containing 1.5% carboxymethylcellulose
(Sigma), 5% (v/v) foetal calf serum and 25mM HEPES buffer.
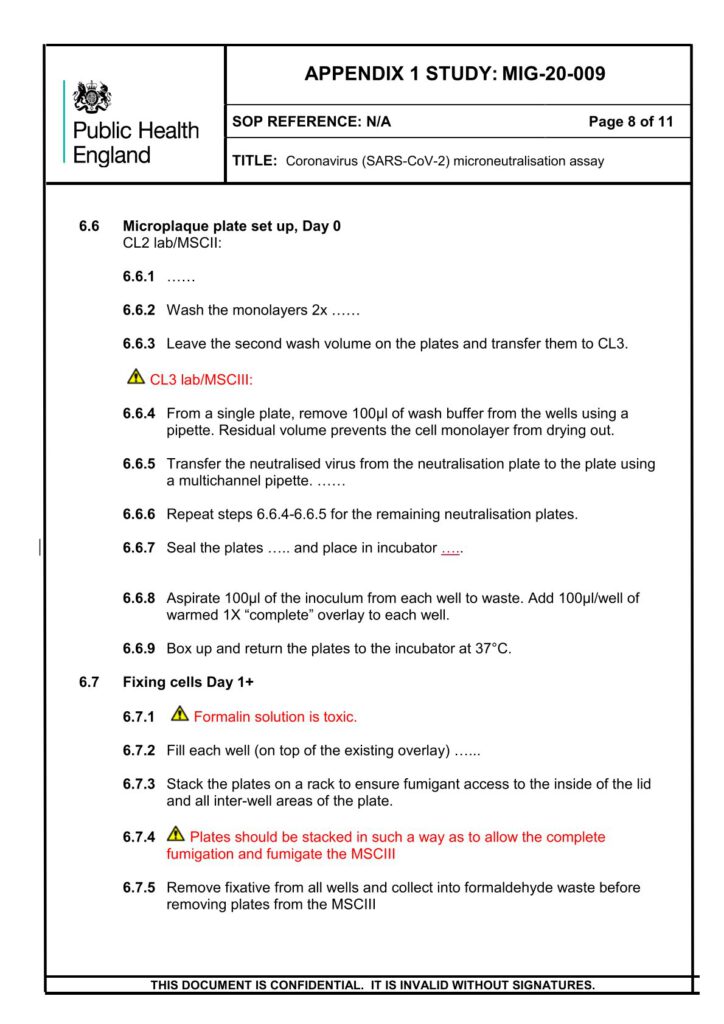
After incubation, at 37°C for 24 hours, plates will be fixed overnight with 20% (v/v) formalin/PBS, washed
with tap water and patches of infected cells (microplaques) detected via an antibody pair specific for the
SARS-CoV-2 RBD Spike protein.
Microplaques are visualized using KPL TrueBlue™ peroxidase substrate and counted on an Immunospot
analyzer. Neutralisation dilution at 50% (ND50) and neutralising factor at 50% (NF50), normalised against
the reference serum, is reported.
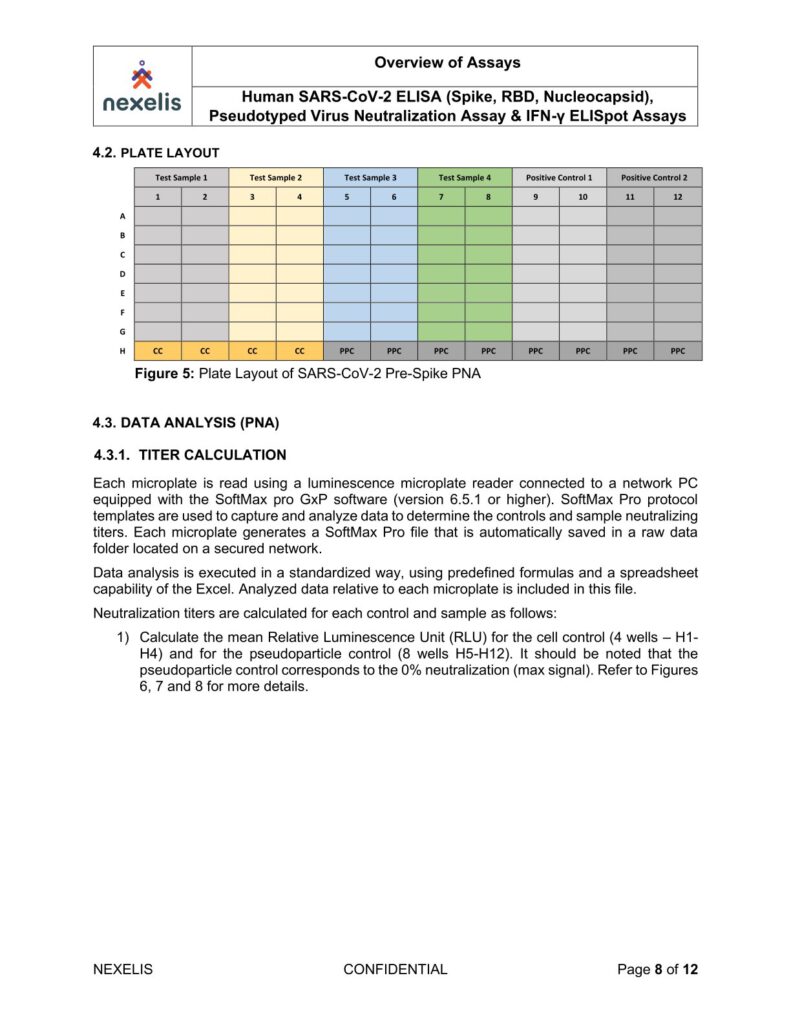
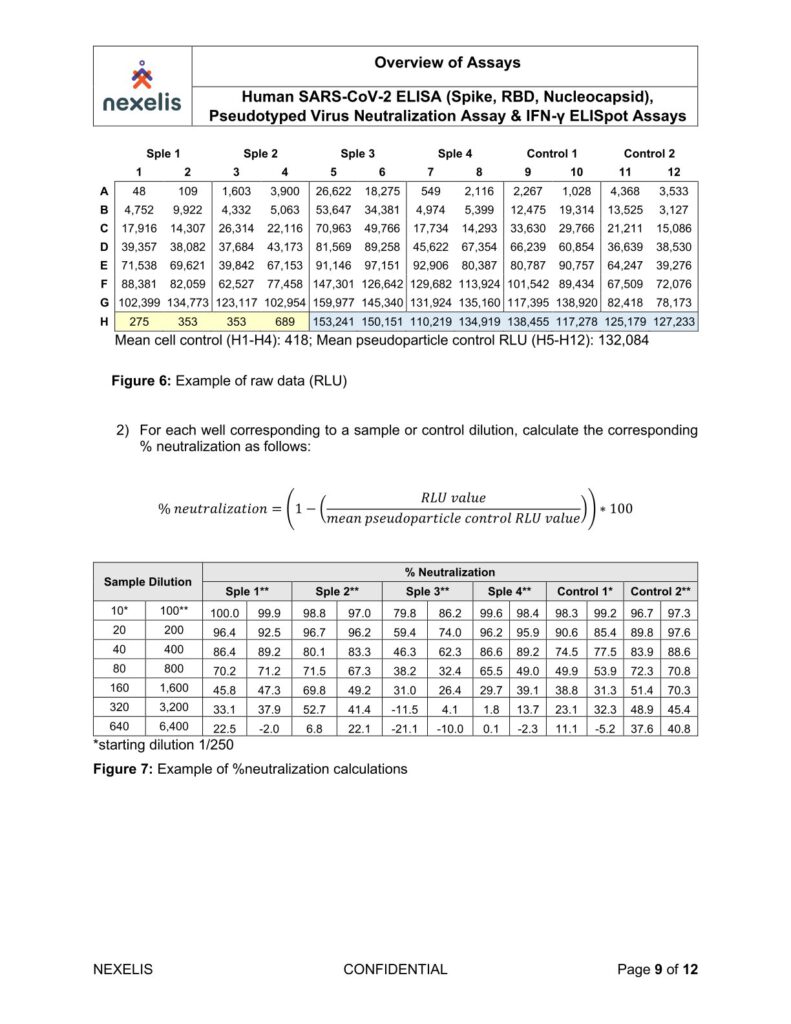
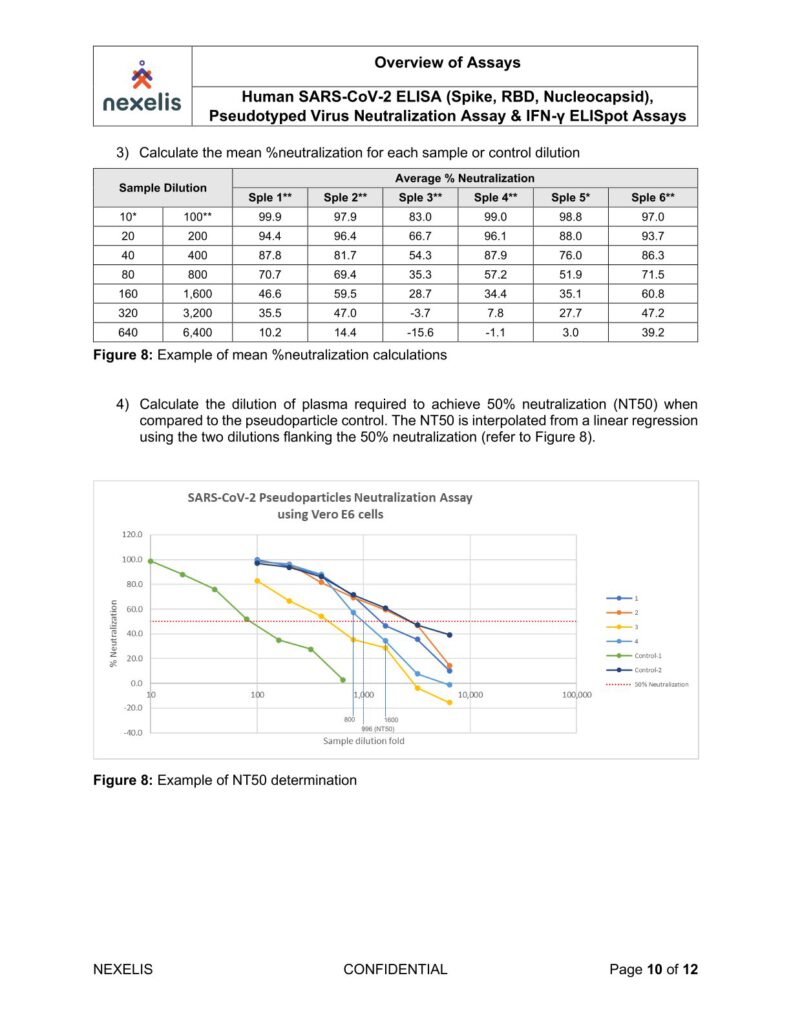
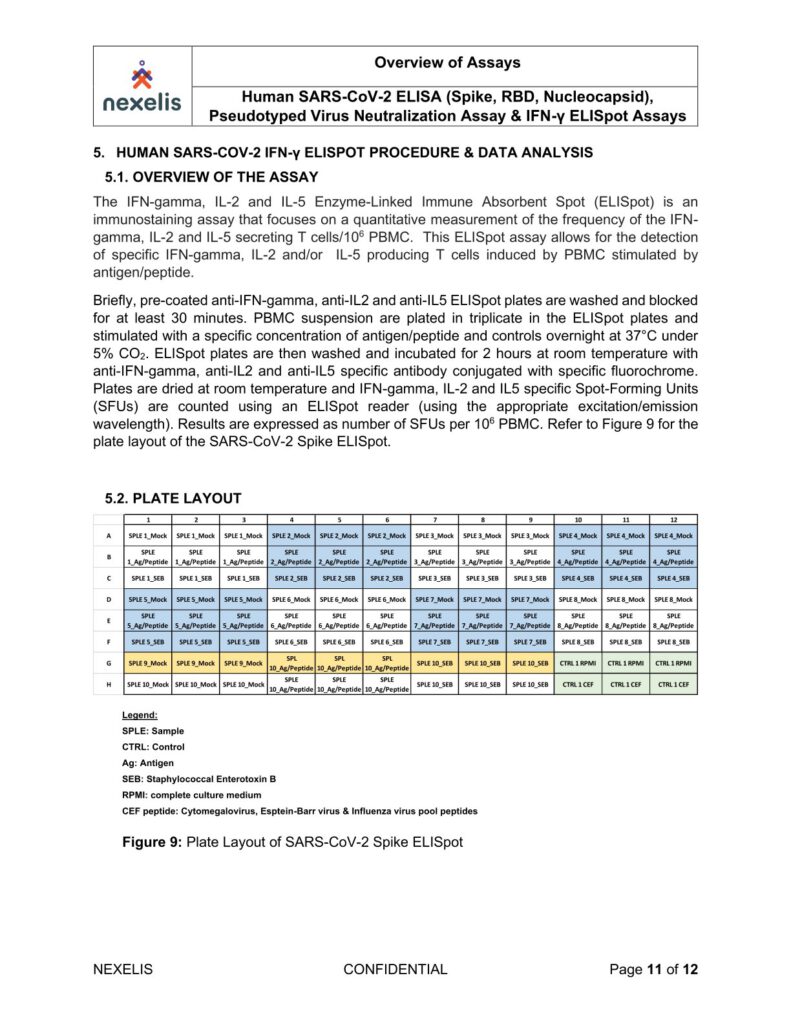
Human SARS-CoV-2 ELISA (Spike, RBD, Nucleocapsid), Pseudotyped Virus Neutralization Assay & IFN-γ ELISpot Assays
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Sed non risus. Suspendisse lectus tortor, dignissim sit amet, adipiscing nec, ultricies sed, dolor.
Work Package Publications
11.2022 / Performance and validation of an adaptable multiplex assay for detection of serologic response to SARS-CoV-2 infection or vaccination (Journal of Immunological Methods): https://www.sciencedirect.com/science/article/pii/S0022175922001326